suppressPackageStartupMessages({
library(dplyr)
library(sf)
library(ggplot2)
library(cmocean)
library(maxnet)
library(stars)
})Visualization
Set up
Load the needed packages.
Set the file location.
here::i_am("tutorial/Steps_visualization.Rmd")here() starts at /Users/eli.holmes/Documents/GitHub/ohw23_proj_marinesdmsLoad the model
We saved this in the previous step. Has the sdm.model, occ.points, abs.points, and env.stars.crop.
fil <- here::here("data", "models", "io-turtle.RData")
load(fil)Load region files. We created our region objects in a separate file Region data and saved these in data/region. We will load these now.
Load the bounding box polygon and create a bounding box.
#Loading bounding box for the area of interest
fil <- here::here("data", "region", "BoundingBox.shp")
extent_polygon <- sf::read_sf(fil)
bbox <- sf::st_bbox(extent_polygon)Predicting
clamp <- TRUE # see ?predict.maxnet for details
type <- "cloglog"
predicted <- predict(sdm.model, env.stars.crop, clamp = clamp, type = type)
predictedstars object with 2 dimensions and 1 attribute
attribute(s):
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
pred 0.0047245 0.1095251 0.1561052 0.1812027 0.2369365 0.9996747 52802
dimension(s):
from to offset delta refsys x/y
x 2663 2942 -180 0.08333 +proj=longlat +datum=WGS8... [x]
y 695 1082 90 -0.08333 +proj=longlat +datum=WGS8... [y]Visualization
ggplot() +
geom_stars(data = predicted) +
scale_fill_cmocean(name = "ice", direction = -1, guide = guide_colorbar(barwidth = 1, barheight = 10, ticks = FALSE, nbin = 1000, frame.colour = "black"), limits = c(0, 1)) +
theme_linedraw() +
coord_equal() +
theme(panel.background = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()) +
labs(title = "Loggerhead and green sea turtle SDM in the Arabian Sea",
x = "Longitude",
y = "Latitude",
fill = "Probability",
shape = "Species (presence)",
subtitle = "Environmental predictors: mean SS temp, mean SS salinity, mean bathymetry, \nmean pH, mean DO, mean SS chlorophyll-a, mean SS nitrate") +
geom_point(occ.points, mapping = aes(shape = common.name, geometry = geometry), stat = "sf_coordinates", alpha = 0.3, color = "purple") +
#scale_x_continuous(breaks = seq(40, 70, 10), limits = c(42, 70))+
scale_y_continuous(breaks = seq(0, 30, 10))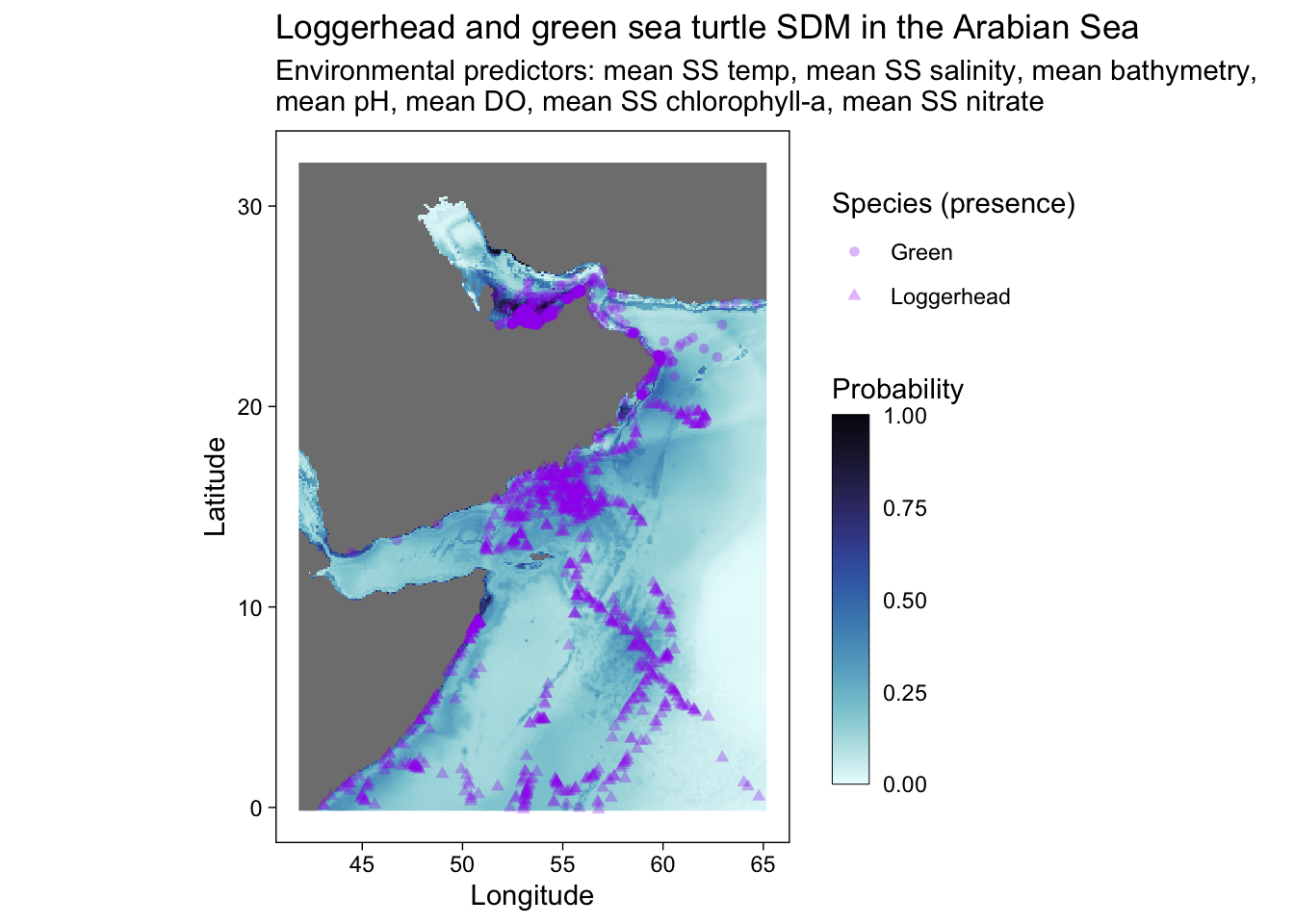
# ggsave("SDM_loggerhead_green_w points.pdf", height = 6, width = 8.5)# ggplot - without occurrence data points
ggplot() +
geom_stars(data = predicted) +
scale_fill_cmocean(name = "ice", direction = -1, guide = guide_colorbar(barwidth = 1, barheight = 10, ticks = FALSE, nbin = 1000, frame.colour = "black"), limits = c(0, 1)) +
theme_linedraw() +
theme(panel.background = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()) +
labs(title = "Loggerhead and green sea turtle SDM in the Arabian Sea",
x = "Longitude",
y = "Latitude",
fill = "Probability",
shape = "Species (presence)",
subtitle = "Environmental predictors: mean SS temp, mean SS salinity, mean bathymetry,\nmean pH, mean DO, mean SS chlorophyll-a, mean SS nitrate") +
#geom_point(occ.points, mapping = aes(shape = common.name, geometry = geometry), stat = "sf_coordinates", alpha = 0.3, color = "purple") +
#scale_x_continuous(breaks = seq(40, 70, 10), limits = c(42, 70))+
scale_y_continuous(breaks = seq(0, 30, 10))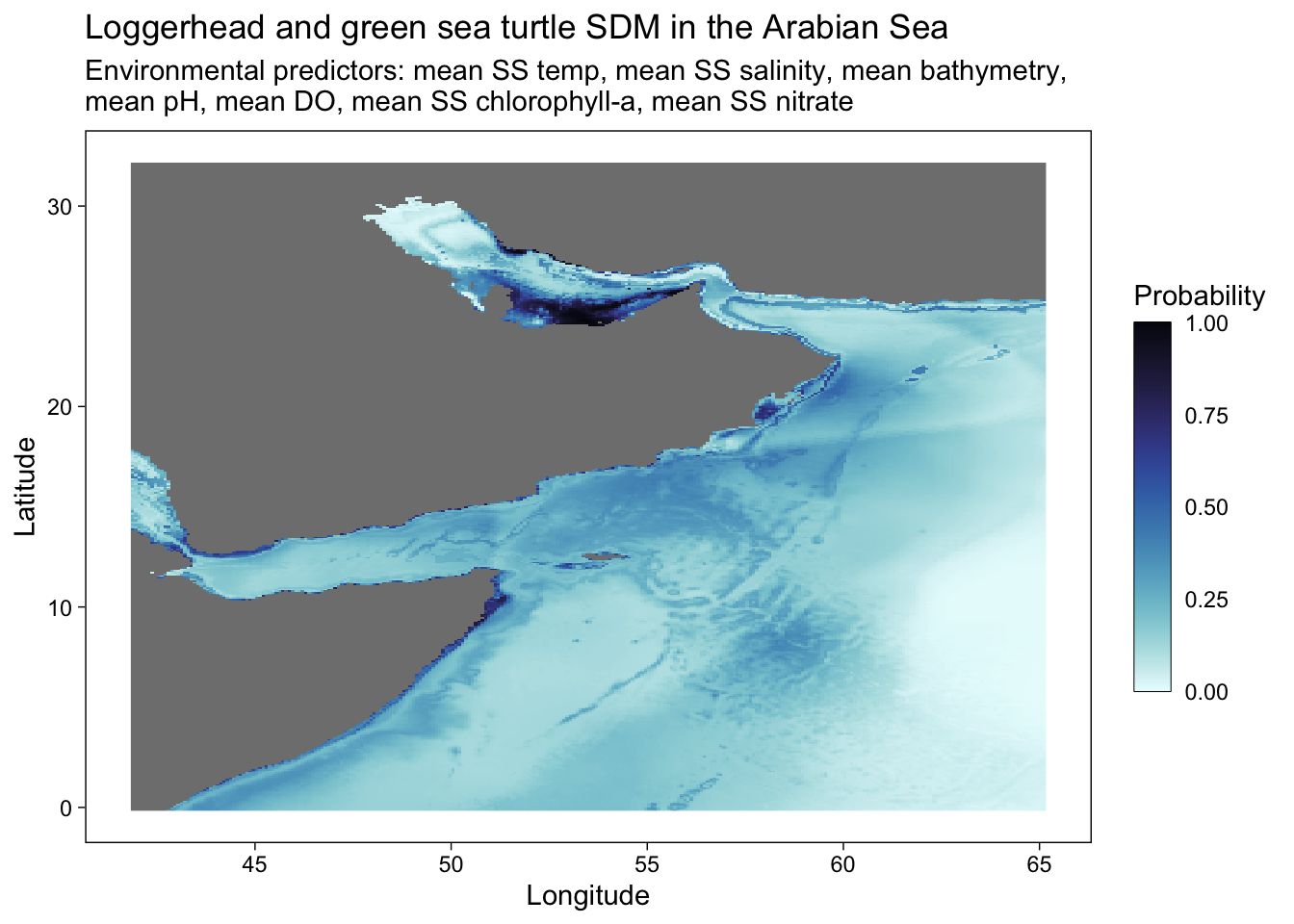
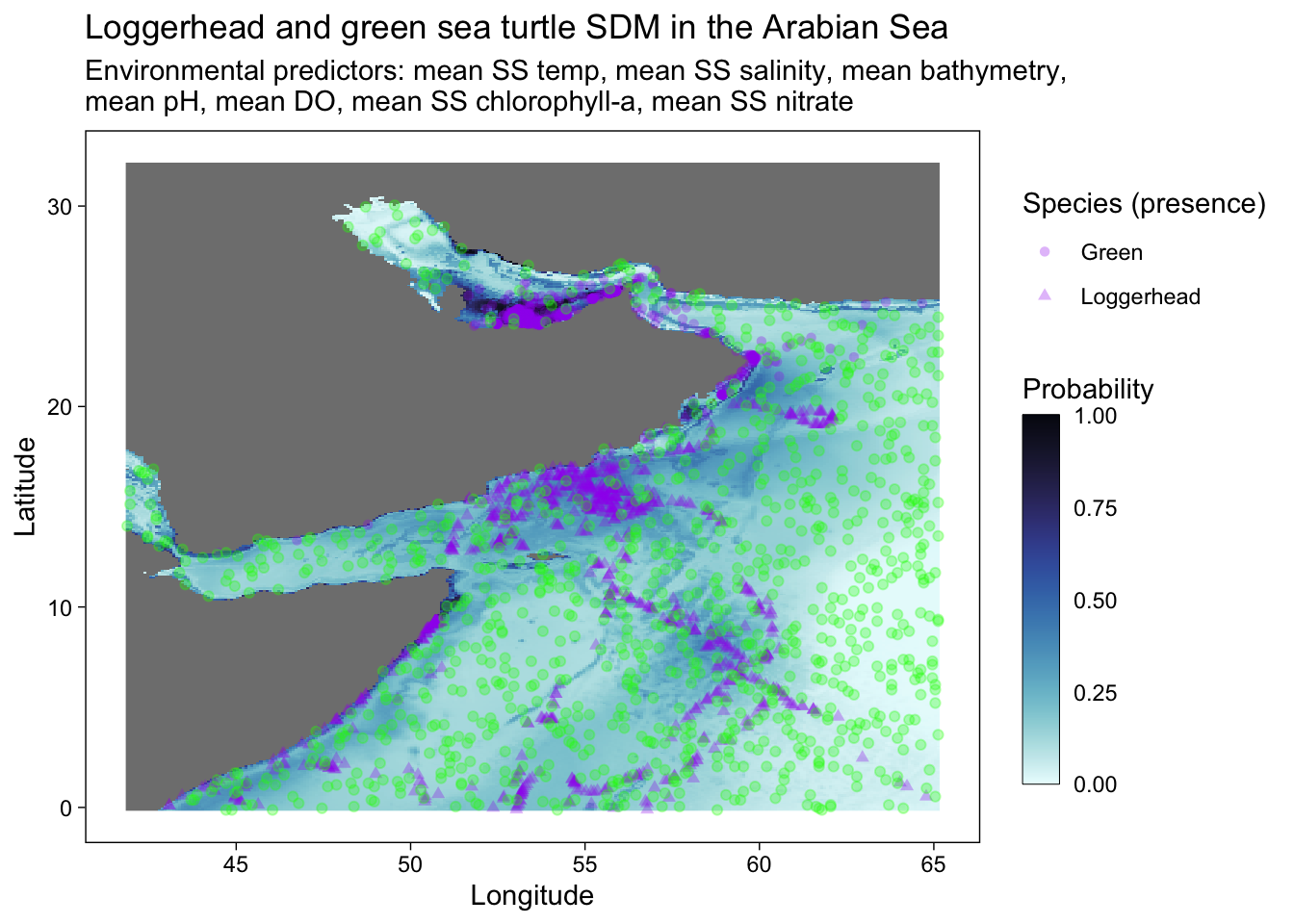
# ggsave("SDM_loggerhead_green.pdf", height = 6, width = 8.5)# ggplot - with occurrence (purple) and absence (green) data points
ggplot() +
geom_stars(data = predicted) +
scale_fill_cmocean(name = "ice", direction = -1, guide = guide_colorbar(barwidth = 1, barheight = 10, ticks = FALSE, nbin = 1000, frame.colour = "black"), limits = c(0, 1)) +
theme_linedraw() +
theme(panel.background = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()) +
labs(title = "Loggerhead and green sea turtle SDM in the Arabian Sea",
x = "Longitude",
y = "Latitude",
fill = "Probability",
shape = "Species (presence)",
subtitle = "Environmental predictors: mean SS temp, mean SS salinity, mean bathymetry, \nmean pH, mean DO, mean SS chlorophyll-a, mean SS nitrate") +
geom_point(occ.points, mapping = aes(shape = common.name, geometry = geometry), stat = "sf_coordinates", alpha = 0.3, color = "purple") +
#scale_x_continuous(breaks = seq(40, 70, 10), limits = c(42, 70))+
scale_y_continuous(breaks = seq(0, 30, 10)) +
geom_point(abs.points, mapping = aes(geometry = geometry), stat = "sf_coordinates", alpha = 0.3, color = "green") # adding in absence data